
Acute myeloid leukemia arising to genetic susceptibility genes related T cell acute lymphoblastic leukemia: case report
Introduction
Fanconi anemia (FA), autosomal recessive or X-linked disease, is heterogeneous phenotype. Associated with abnormal response to DNA damage, FA is characterized by congenital abnormally, progressive bone marrow failure, and predisposition to neoplasm (1). There was 16% of FA patients developed hematologic malignancies and 10% of them developed solid tumors (2). While Myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) are the most common hematologic malignancies seen in patients with FA, cases of acute lymphoblastic leukemia (ALL) have also been described in the literature (3). Homozygous or compound heterozygous mutations are usually detected in patients with FA (1), hence we reported a rare case with FA who presented with precursor T cell ALL (T-ALL) at onset, secondary AML arose after he received chemotherapy and got complete remission (CR), however inherited heterozygote mutation of FANCC and heterozygous nonsynonymous single nucleotide variability (SNV) mutation of AKAP9 were confirmed.
We present the following case in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/acr-20-151).
Case presentation
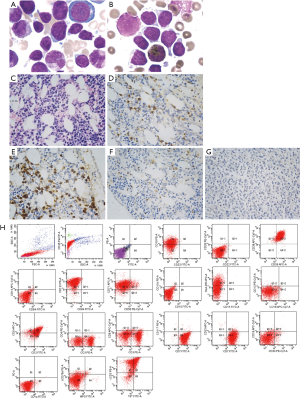
A 12 years 5 months old boy was admitted with fever, pale, hepatosplenomegaly, lymphadenectasis and pancytopenia. He had no growth retardation, past history and family history of tumor or cytopenia were also not mentioned. Initial complete blood count (CBC) showed as: white blood cell (WBC) count 1.7×109/L, red blood cell (RBC) count 2.83×1012/L, hemoglobin (Hgb) 84 g/L, platelet (PLT) count 62×109/L, blast cell was not detected in peripheral blood smear. FAB subtype, which was checked by bone marrow (BM) smear (Figure 1A,B) and biopsy (Figure 1C,D,E,F,G), was ALL, evidence of myelodysplasia were not found; immunophenotype of T-ALL was confirmed by immunohistochemical staining (Figure 1A) and flow cytometry, positive myeloid biomarkers were undetectable (FCM, Figure 1H); chromosome karyotype is normal (46,XY[20]); fluorescence in situ hybridization (FISH) of ETV6/RUNX1, TCF3/PBX1, BCR/ABL and MLL rearrangement (MLLr) were negative; common fusion genes were screened by multiplex nest reverse transcriptase polymerase chain reaction (multiplex nested RT-PCR) as literature report (4) and results were also negative. Radiographs of the whole skeleton were normal. The patient received chemotherapy according to modified St. Jude Total XV protocol for newly diagnosed pediatric ALL (5). He achieved CR after a course of induction remission, but severe cytopenia (Lowest value of WBC or absolute neutrophils count was 0.34×109/L or 0.02×109/L) was presented, sepsis and invasive fungal infection arose. He was classified as intermediate risk (IR) group by minimal residential disease (MRD) monitoring as protocol request, chemotherapy was followed by a course of consolidation, continuous and maintenance therapy (5).
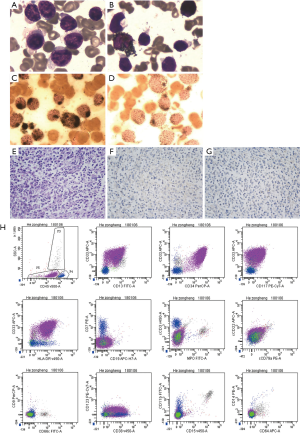
Seventeen months later (in course of maintenance therapy), the patient was again presented with fever, pallor, osteodynia and hepatosplenomegaly. CBC showed as: WBC 27.39×109/L with 85% blast cell, RBC 1.88×1012/L, Hgb 61 g/L, PLT 73×109/L. FAB subtype was classified as AML with myelodysplasia by BM smear (Figure 2A,B,C,D) and biopsy (Figure 2E,F,G), AML was also confirmed by FCM (Figure 2H); chromosome karyotype was normal; FISH of RUNX1/ETO, CBFb, PML/RARa, BCR/ABL and MLLr and fusion genes screen as literature (4) were negative.
BM sample of the patient in ALL status, which was preserved in liquid nitrogen, and in AML status, peripheral blood samples of his elder sister and parents were collected, whole exome sequencing (WES) was detected for these samples by next generation sequencing (NGS), detectable pathologic gene mutations were checked by Sanger sequencing (6). All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). The parents of the patient provided written informed consent to publish the case report and accompanying images.
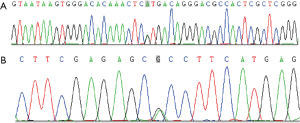
Somatic pathologic mutations of WT1, DNM2, GATA and JAK3 were found in the AML status and ALL status samples. Heterozygous nonsense mutation of FANCC (NM_001243744:exon7:c.C553T: p.R185X) and heterozygous nonsynonymous mutation of AKAP9 (NM_147185:exon23:c.G5705A: p. R1902H) were detected in the sample of the patient under both ALL and AML status; heterozygous mutation of FANCC and AKAP9 was detected in his father and mother respectively, while heterozygous mutation of FANCC was also detected in his elder sister (Figure 3). Peripheral blood lymphocytes of the patient in 1st CR status which was preserved in liquid nitrogen were collected, mitomycin C (MMC) induced chromosomal breakage tests were done as literature reports for the family (7,8), positive results was found in the patient (Figure 4) and negative results were detected in his parents and sister.
Accumulate dosage of DNR was 210 mg/m2 in preceding treatment for ALL, therefore the patient received chemotherapy consisted of etoposide (VP-16 100 mg/m2/d, day1–3) and cytarabine (AraC 200 mg/m2/d, day1–7). Severe side effects, including significant myelosuppression and severe infection, were presented; the patient died of sepsis and invasive fungal infection three weeks after chemotherapy.
Discussion
FA is divided into several subgroups according to its genetic mutations (1). Sixteen known mutated genes that are associated with FA had been reported, most of such subgroups are autosomal recessive diseases, of which FANCB mutation is X-linked disease (9). Patients with FA are characterized by hypersensitivity to DNA cross-linking or alkylating agents, such as ultra-violet radiation and MMC, that block DNA replication and RNA transcription, chromosomal breakage test and comet assay test play important role in the diagnosis of FA (8,9). Homozygous or bilateral heterozygote mutations are usually detected in FA patients (1), however heterozygote mutation of FANCC and heterozygous nonsynonymous SNV mutation of AKAP9 were confirmed in the reported patient; clinical diagnosis of FA was considered because of positive results for chromosomal breakage test and comet assay test.
Associated with abnormal response to DNA damage, FA patients have a very high risk of congenital malformation, bone marrow failure and malignancies (2). Although FA is well-known for its relationship to hematologic malignancies, literature reviews had suggested that FA patients are highly predisposed to solid tumors (1). A review of 1301 FA patients who were diagnosed from 1927 to 2001 had reported 192 (14.76%) cases of leukemia/MDS and 105 (8.07%) cases of solid tumor (3); 754 FA patients were enrolled in a 20-year prospective study on the International Fanconi Anemia Registry, 601 cases (79.71%) suffered from bone marrow failure, 173 cases (22.94%) suffered from neoplasms which included 120 cases of hematologic malignancies and 79 cases of non-hematologic neoplasm respectively (2). The most common subgroup of hematologic malignancies in FA patients is AML/MDS of which incidence is approximately 15%, whereas ALL, in which most of them were T-ALL, is rare (10,11).
In literature reports (1,8,9), FA patients presented with homozygous or bilateral heterozygosity mutation, whereas heterozygosity mutation of FANCC and AKAP9 has not reported in FA patients. The A-kinase anchor proteins (AKAPs) are a group of structurally diverse proteins, which have the common function of binding to the regulatory subunit of protein kinase A (PKA) and confining the holoenzyme to discrete locations within the cell (11). There are at least 50 members, often named after their molecular mass, and AKAP9 protein is encoded by AKAP9 gene which is located in 7q21.2. AKAPs are widely distributed in many kinds of cells. Different signal transduction enzymes are recruited near the substrate. Thus, upstream activators and downstream factors are effectively assembled into a macromolecular signal transduction complex, which integrates various signal molecules that co-regulate the phosphorylation of cell substrates, thus ensuring the specificity and accuracy of signal transduction (12). We speculate that AKAP9 protein may interfere with the normal signal transduction of heterozygous mutation expression of FANCC gene and result in inactivation of FANCC gene function. Unfortunately, the patient died of sepsis and we don’t have enough blood samples to explore the role of AKAP9 mutation in patients with heterozygosity FANCC mutation.
Conclusions
In brief, ALL in patients with FA is rarely reported, we present a FA patient who was lack of classic FA manifestation and FA family history, he was onset with T-ALL and secondary AML with myelodysplasia was developed after he achieved CR. Literature reports about ALL related to FA patients are limited and most of such patients were T-ALL. Primary diagnosis of FA usually is ignored when patients presented with ALL, WES should be detected when the secondary tumor arose; further studies are needed to determine for the appropriate chemotherapy regimen in these patients.
Most FA patients were detected with homozygous or bilateral heterozygote mutation, the reported patients were detected heterozygote mutation of FANCC and heterozygous nonsynonymous SNV mutation of AKAP9, we suspected that AKAP9 gene and its protein affect FANCC gene by interfering with signal transduction, which leads to the occurrence and development of leukemia, further research is needed to confirm it.
Acknowledgments
Funding: The study was supported by the National Natural Science Foundation of China (Project No. 81900162) and the Chongqing Science and Technology Commission of China PR (Project No. cstc2018jsyj-jsyjX0015).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/acr-20-151
Peer Review File: Available at http://dx.doi.org/10.21037/acr-20-151
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/acr-20-151). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval for the publication of this case was obtained from the ethical committee of Children Hospital of Chongqing Medical University. All procedures performed in study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). The parents of the patient provided written informed consent to publish the case report and accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bagby GC, Alter BP. Fanconi anemia. Semin Hematol 2006;43:147-56. [Crossref] [PubMed]
- Kutler DI, Singh B, Satagopan J, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 2003;101:1249-56. [Crossref] [PubMed]
- Alter BP. Cancer in Fanconi anemia, 1927-2001. Cancer 2003;97:425-40. [Crossref] [PubMed]
- Pallisgaard N, Hokland P, Riishøj DC, et al. Multiplex reverse transcription-polymerase chain reaction for simultaneous screening of 29 translocations and chromosomal aberrations in acute leukemia. Blood 1998;92:574-88. [Crossref] [PubMed]
- Karol SE, Yang W, Van Driest SL, et al. Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood 2015;126:1770-6. [Crossref] [PubMed]
- Spinella JF, Healy J, Saillour V, et al. Whole-exome sequencing of a rare case of familial childhood acute lymphoblastic leukemia reveals putative predisposing mutations in Fanconi anemia genes. BMC Cancer 2015;15:539. [Crossref] [PubMed]
- Solanki A, Mohanty P, Shukla P, et al. FANCA Gene Mutations with 8 Novel Molecular Changes in Indian Fanconi Anemia Patients. PLoS One 2016;11:e0147016 [Crossref] [PubMed]
- Howlett NG, Taniguchi T, Durkin SG, et al. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 2005;14:693-701. [Crossref] [PubMed]
- Ayas M, Saber W, Davies SM, et al. Precursor T acute lymphoblastic leukemia from myelodysplastic syndrome in Fanconi anemia. J Hematop 2013;3:161-5.
- Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet 2007;44:1-9. [Crossref] [PubMed]
- Herter JM, Grabie N, Cullere X, et al. AKAP9 regulates activation-induced retention of T lymphocytes at sites of inflammation. Nat Commun 2015;6:10182. [Crossref] [PubMed]
- Ikezu T, Chen C, DeLeo AM, et al. Tau Phosphorylation is Impacted by Rare AKAP9 Mutations Associated with Alzheimer Disease in African Americans. J Neuroimmune Pharmacol 2018;13:254-64. [Crossref] [PubMed]
Cite this article as: Guo Y, Liao X, Zou P, Xiao J. Acute myeloid leukemia arising to genetic susceptibility genes related T cell acute lymphoblastic leukemia: case report. AME Case Rep 2021;5:29.



